IGV: A quick overview

The Integrative Genome Browser (IGV) from the Broad Institute can be considered the microscope for the Bioinformatician.
Get IGV
IGV can be easily downloaded from the Broad Institute website.
It’s a Java application with a graphical user interface (GUI), and its concepts revolves around:
- Loading a reference sequence
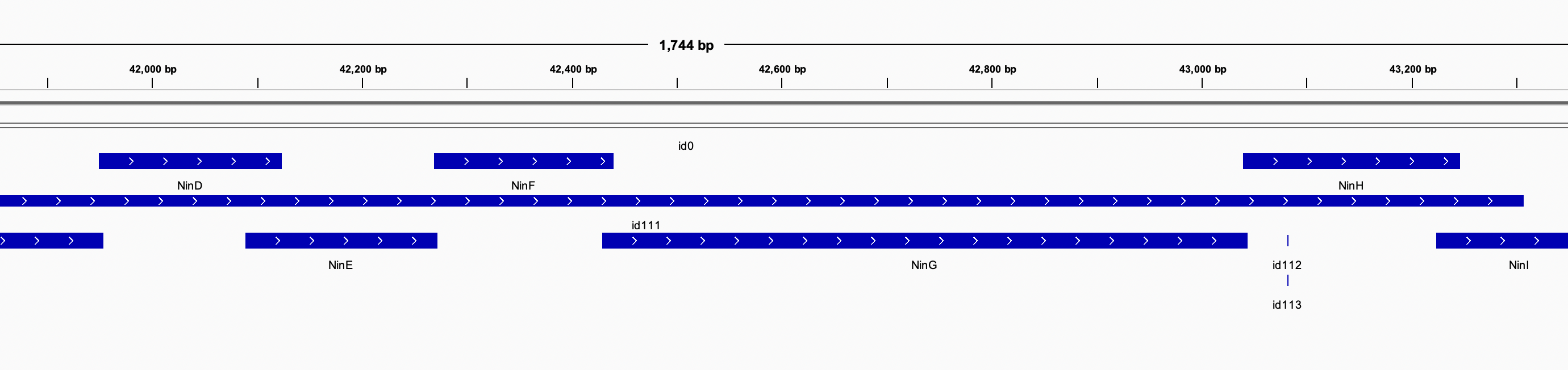
- Adding tracks related to the reference sequence, such as:
- Alignments (BAM files)
- Genomic features, annotations (GFF, GTF, BED files)
- Variants (VCF files)
Load a reference sequence
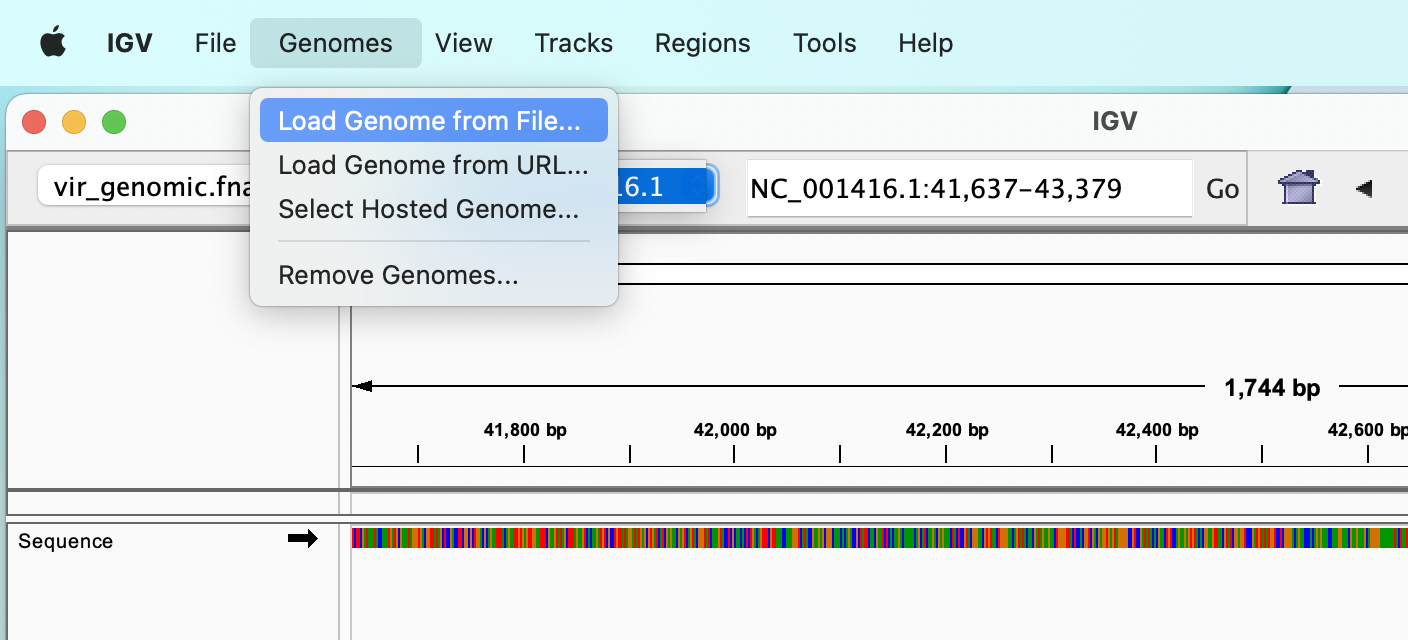
The reference genome can be selected from a drop-down menu, which allows to select one of the many model organisms available, or we can load a FASTA file from our disk, using the “Genomes” :right_arrow: “Load genome from file…” menu.
Load some tracks
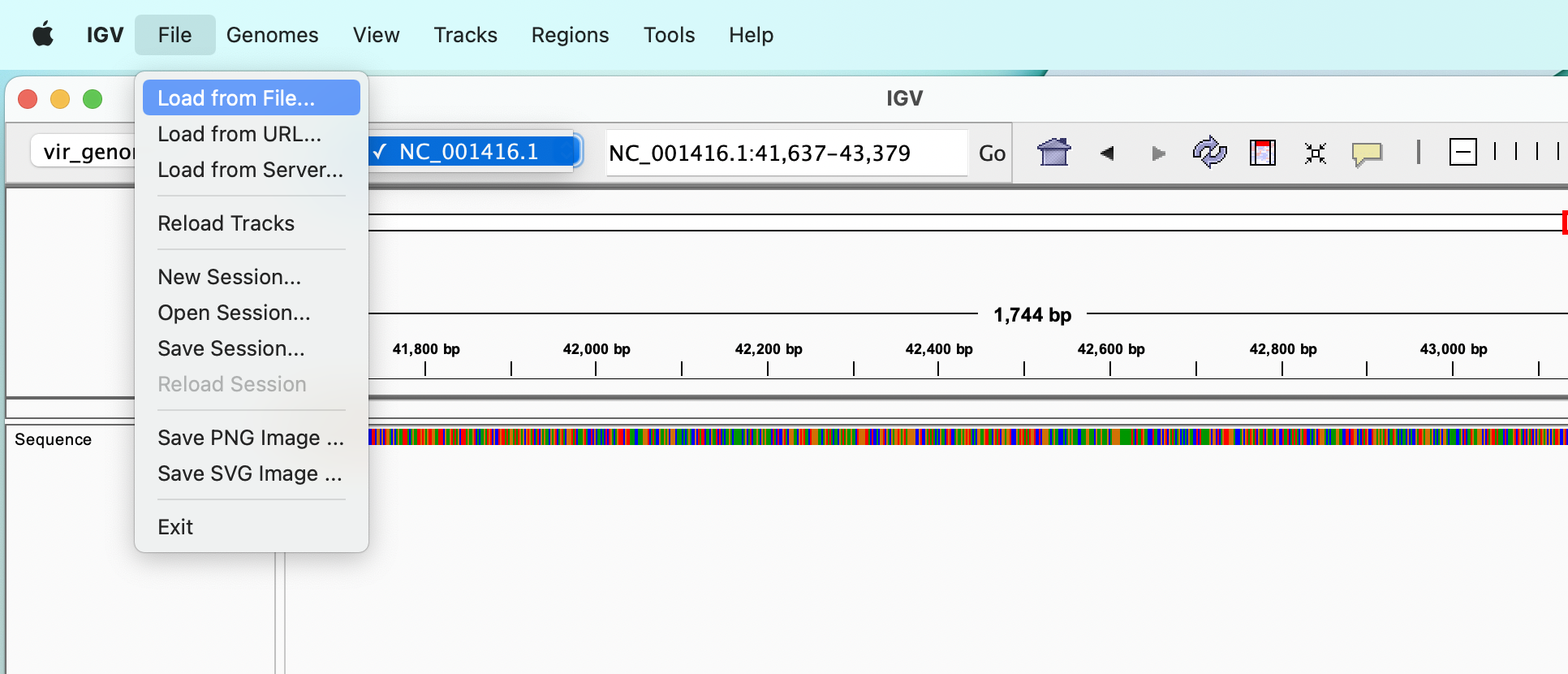
If we have sorted and indexed BAM files, VCF files or annotation files we can load them from the “File” :right_arrow: “Load from File…” menu.
We will see the results plotted.