This is ScaMPI!
During my PhD, I had the privilege to work with Elisa Corteggiani. She started studing the genetic and biochemical properties of Nannochloropsis gaditana, a promising source of biofuel (at least at the time, circa 2010), and planned to do the whole genome sequencing and assembly.
Both 454 FLX libraries and long mate paired libraries (SOLiD) were sequences, but the lack of a scaffolding program properly supporting color space mate paired libraries was a problem. So I wrote ScaMPI, a suite of tools to automatically and manually (!) scaffold the genome.
- To read more, check out my PhD thesis.
- Code is in the ScaMPI repository, for archival purposes only.
Web interface
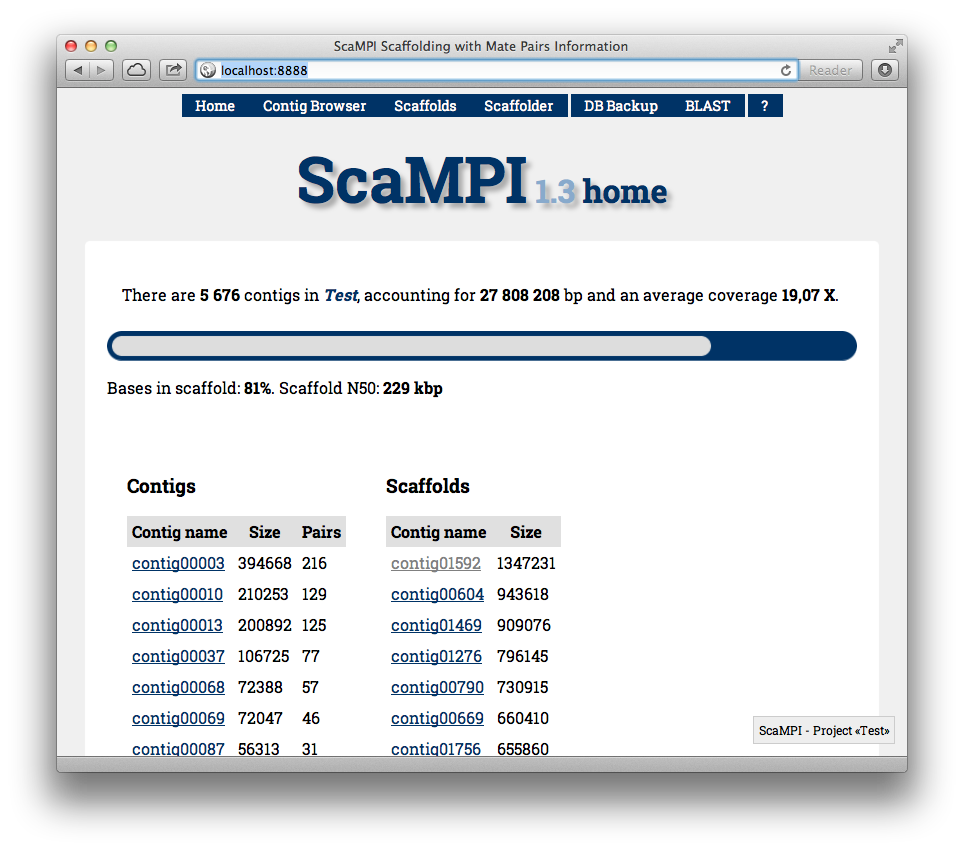
Home page of the ScaMPI web interface.
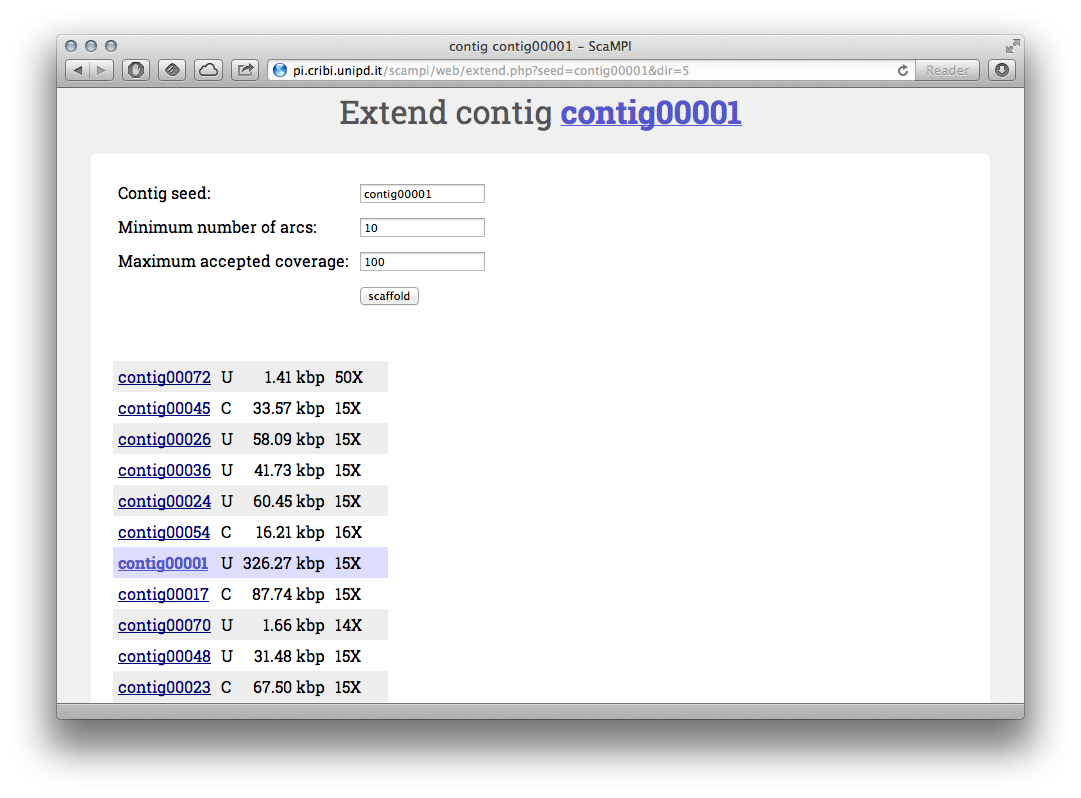
Extension of a “seed” contig, the output will include the orientation listed as C (complemented) or U (uncomplemented).
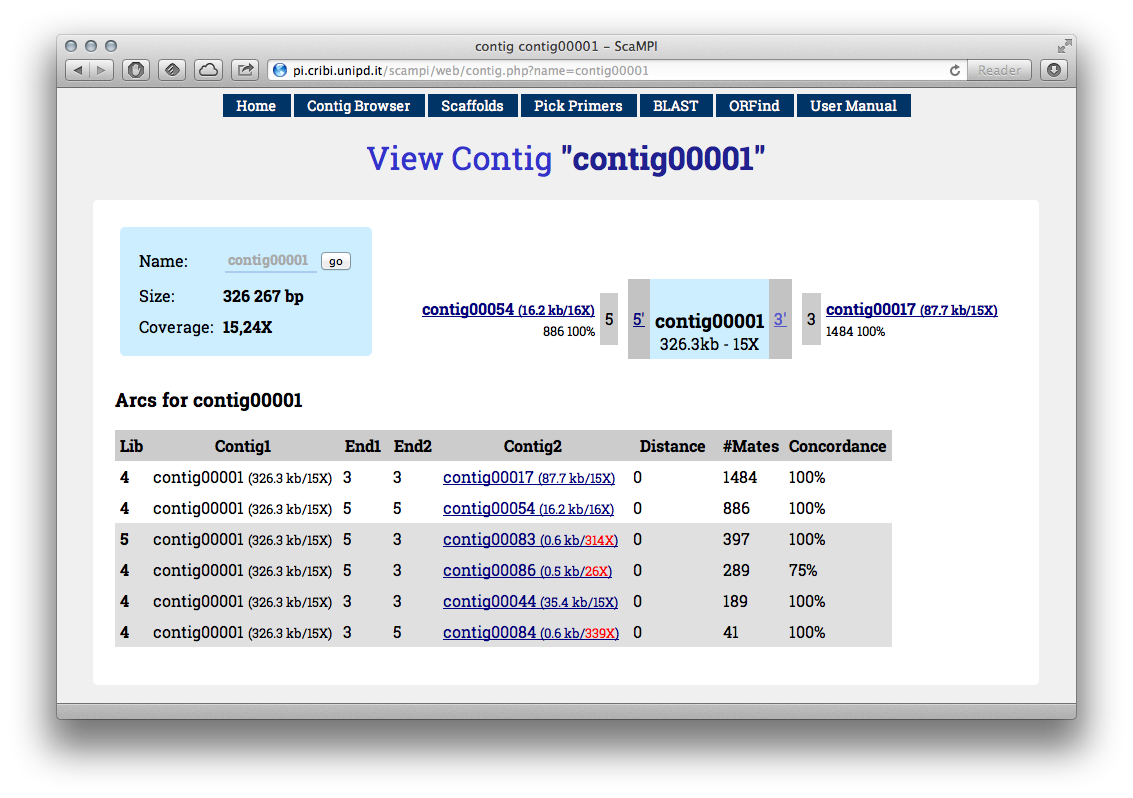
View of a single contig. All the possible connections are listed below, but the flanking contigs are the reasonable possibilities (noise is filtered)