The Ideal Rootstock: A Quest for Sustainable Viticulture
The AGER-SERRES project represents a significant milestone in viticulture research, bringing together multiple Italian research institutions in a collaborative effort to develop next-generation grapevine rootstocks. Launched in 2011, this pioneering initiative focuses on selecting new rootstocks that are resistant to abiotic stresses while promoting sustainable viticulture practices. The project aims to address critical challenges facing modern vineyard management, including water scarcity, climate change adaptation, and the need to reduce chemical inputs. As Professor Attilio Scienza, a key figure in the project, emphasizes, “roots are the brain of the plant and developing improved rootstocks is fundamental for achieving truly sustainable viticulture”.
Through advanced genomic analysis and field testing, SERRES seeks to create rootstocks that can thrive with minimal irrigation and fertilization while maintaining optimal grape quality - essentially working towards what could be considered the “ideal rootstock” for modern viticulture.
In the last decade, genomic analysis technologies have made incredible progress, enabling highly accurate genome sequencing of many organisms, including the human genome (published in 2001) and the grapevine genome (published in 2007). While knowing a genome’s sequence is important, it’s crucial to understand that the sequence alone doesn’t reveal the full meaning of the information it contains.
Genomics and post-genomics
Just as with the human genome, post-genomic research for other organisms involves trying to associate genes with their biological processes to determine each gene’s biological function. However, before this can happen, genes must be identified by locating their physical boundaries within the genomic sequence: start, end, and splicing sites.
In reality, the three processes of sequencing, prediction, and annotation of a genome are never truly complete:
- Sequencing should lead to increasingly precise definition of each species’ reference genome
- Gene prediction continuously improves through cDNA analysis and RNA sequencing
- Gene annotation evolves with functional genomics research
The M1 and M4 genotypes
Among the key achievements of the AGER-SERRES project was the development and characterization of several promising rootstock genotypes, with M1 and M4 emerging as particularly noteworthy candidates. M1, derived from a cross between 106/8 (V. riparia × (V. cordifolia × V. rupestris)) and Resseguier n.1 (V. berlandieri), demonstrates high grafting compatibility, reduced vigor, and excellent resistance to iron chlorosis, while maintaining moderate salt tolerance. M4, created by crossing 41B (V. vinifera × V. berlandieri) with Resseguier n.1, stands out for its exceptional drought resistance and high salt tolerance, combined with moderate vigor.
Both genotypes represent significant advances in rootstock breeding, offering improved adaptability to environmental stresses while maintaining high grafting success rates - crucial characteristics for modern viticulture facing climate change challenges. Field trials across multiple sites have demonstrated that these rootstocks not only perform well under stress conditions but also enhance the qualitative characteristics of the grafted varieties.
Rootstock Genome Sequencing
The first grapevine genome sequence was published in Nature in 2007, based on a highly homozygous Pinot Noir genotype called PN40024. This identified approximately 30,000 genes, of which about 26,000 were annotated. The PN40024 sequence has since served as a crucial reference point for all genetic studies in grapevine.
Within the SERRES project, the genomics research group led by Prof. Giorgio Valle sequenced and assembled the genomic sequence of the new genotypes (M1, M4 and their parental 101.14). I was working as bioinformatician in the project, leading the assembly and variant calling parts.
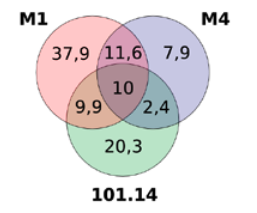 Figure 1: Distribution of genetic variants among three rootstock genotypes (M1, M4, and 101.14) compared to the reference genome. The Venn diagram shows the percentage of variants shared between genotypes. Only 10% of variants are common to all three genotypes.
Figure 1: Distribution of genetic variants among three rootstock genotypes (M1, M4, and 101.14) compared to the reference genome. The Venn diagram shows the percentage of variants shared between genotypes. Only 10% of variants are common to all three genotypes.
| Rootstock | High-quality variants | SNPs | InDels | Density (variants per 2Kb) |
|---|---|---|---|---|
| M4 | 1,671,472 | 1,654,955 | 16,517 | 7.89 ± 7.17 |
| M1 | 3,641,652 | 3,585,346 | 56,306 | 15.63 ± 10.73 |
| 101.14 | 2,233,051 | 2,186,705 | 46,346 | 9.72 ± 7.11 |
Conclusions
The genomic analysis of the three rootstocks provided interesting and partly unexpected results:
-
Rootstock properties appear to be based on polymorphisms rather than unique genes - they share essentially the same genes as V. vinifera but with different variants.
-
Surprisingly, the differences between rootstocks and the PN40024 reference genome are similar in magnitude to those found within V. vinifera species, despite rootstocks being derived from crosses between different Vitis species.
-
The similarity between rootstock genomes and PN40024 sequence enabled the use of AGER-SERRES project data to significantly improve gene prediction and annotation of the V. vinifera reference genome.
Practical Applications
This research contributes significantly to understanding the molecular basis of genotype/phenotype relationships in grapevine rootstocks, providing a platform for associating genes with phenotypes and exploiting natural genetic diversity to develop superior genotypes.
A summary of the book chapter Il genoma dei portainnesti: similarità e differenze con Vitis vinifera, by Valle, Vitulo, Vannozzi, Telatin and Lucchini